We are all genetic mutants
If you are like most people, during the course of your life you have rarely, if ever, thought about your genes. Maybe you found some third cousins or suspicious half-brothers using a genetic genealogy service, or maybe you did genetic testing during pregnancy that told you the sex of your baby.
Even if you've thought a bit about your genetics, though, you probably couldn't name any specific genetic variants in your genome. And why should you? There's not much you can do with that information anyways.
Over the past few years, a handful of trends have come together that will make it essential for every health-focused individual to know the handful of most important genetic variants in their genome – if you're the type of person who tests your LDL cholesterol every year or two, you'll also want to check for updates in our understanding of your unique genetic profile.
These two trends are in two buckets – advances in understanding your genome and taking action:
- Understanding your genome: Large genome sequencing studies are showing that nearly everyone has a set of rare, high-impact genetic variants that influence a range of noticeable traits, from blood measurements to body composition to behavior.
- Taking action: Changes in molecular biology technologies now allow you to make decisions at different points in your life based on your genetic variants – DNA sequencing from embryos means you can choose which variants to pass on to your children, gene therapies mean you may be able to alter some of your own genetic variants, and so on.
Let's dig in to each of these separately.
Understanding your genome
Large-scale genome sequencing in cohorts like the UK Biobank and All Of Us have made fundamental breakthroughs in understanding the specific genetic variants that make you unique. There is now a steady stream of discovery of large-effect, rare variants on a wide range of traits.
The understanding and implication of these variants has barely registered in academia, much less out in the general public. To make this concrete, let's take a few examples:
1. One in 3,000 people have a KDM5B mutation that causes a sub-clinical intellectual disability syndrome. This comes from a paper using the UK Biobank from 2023. Mutations in the gene KDM5B are known to cause severe intellectual disability in a recessive form; what has come from the UK Biobank is that relatively rare (but not that rare) carriers of mutations in the gene have on average around 5-10 points lower IQ, lower hand grip strength, increased risk of atrial fibrillation, and generally what seems like a more 'mild' version of the recessive disorder, probably out of sight of the medical system.
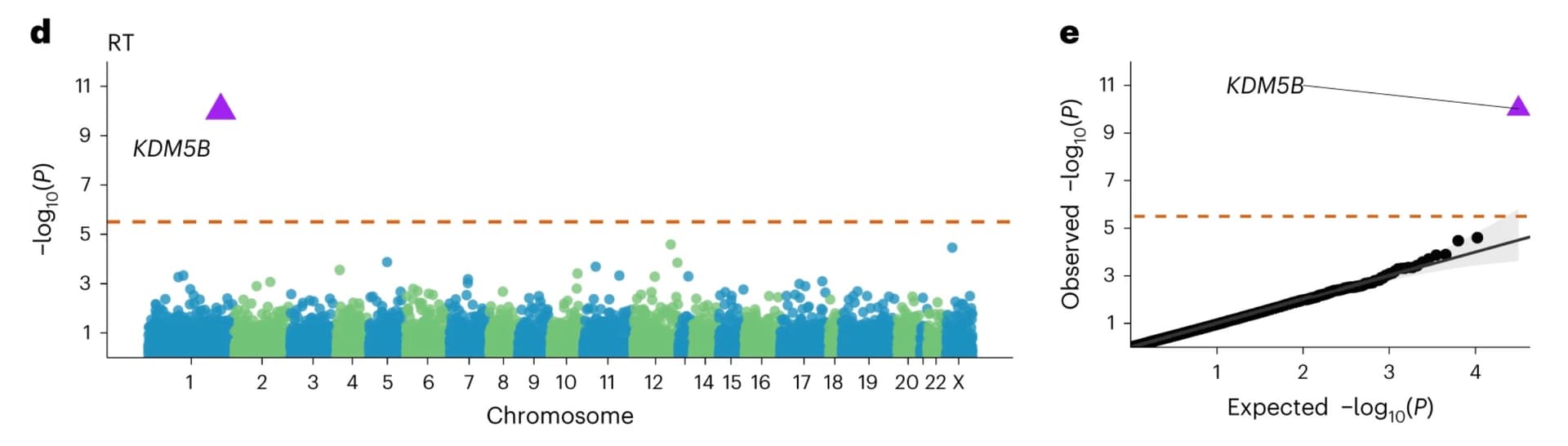
Association study of protein-truncating variants with reaction time, from Chen et al.
In a world where you want to take action to manage and delay the physical/mental declines of aging, knowing a baseline fact like if you have this variant seems like table stakes.
Note I could write similar paragraphs about loss-of-function variants in ANKRD12 (one in 2,500 people) or GIGFY1 (one in 5,000 people).
2. One in 5,000 people have a PER2 loss-of-function mutation that shifts their average sleep cycle one hour earlier. This comes from a paper published on the UK Biobank in 2022. The gene PER2 is a well-known regulator of circadian rhythms, and some genetic variants in the gene have long been known to shift sleep cycles by a few minutes. What has come from the UK Biobank is again that relatively rare (but again not that rare) loss-of-function mutations have a huge effect – a variant with an biological effect of going to sleep and waking up an hour earlier on average must feel absolutely enormous to the person with the variant (given that, as we all know, there are external pressures like jobs/children that result in sleep cycles being only partially under our control).
In a world where you are tracking and optimizing your sleep schedule to maintain mental and physical acuity, knowing if you have this variant again seems like table stakes.
Note that loss-of-function variants in PER3 have similar effects and are present in around one in 1,500 people.
3. One in 10,000 people are unaware that they have a PKD2 mutation that causes progressive decline in kidney function and increased liver and heart failure. This comes from a preprint on the All Of Us cohort from earlier this year. Dominant PKD2 mutations are a well-known cause of polycystic kidney disease; what this paper shows is that for about half of the individuals with a pathogenic mutation in the disease, there's no corresponding diagnosis in the individual's medical records. This could be because these individuals have a somewhat less severe version of the syndrome, and have simply not experienced severe-enough symptoms to interact with the medical system.

A large fraction of carriers of dominant disease mutations have no corresponding diagnosis, from Felker et al.
Common early symptoms of PKD2 mutations include chronic back pain and headaches – in a world where you are spending time and money on managing these types of symptoms, it seems like knowing that the cause is progressive kidney disease would be useful.
Similar results on rates of undiagnosed and likely sub-clinical carriers hold for PKD1 (one in 3,000 people), NF1 (one in 2,000 people), and FBN1 (one in 3,000 people).
If you add up the frequencies of the variants mentioned above, you'll start to wonder – how many variants like this are there? Is it likely that I have one of them? I've now curated variants from the literature and just over 400 genes have rare variants with large effects in these cohorts. At an average frequency in the range of about one in a thousand people, around 40% of the population has a variant known today to have relatively high impact.
And of course biobank studies today include 'only' hundreds of thousands of people – once they successfully reach millions of people another tranche of variants will be uncovered, and once they hit tens of millions of people another tranche again. The picture that is emerging is that everyone has a set of rare, high-impact variants that are noticeable to them in their daily lives.
Taking action
Of course, knowing something is not particularly interesting if you can't actually do anything with the information. Most of these variants are not 'actionable' in the traditional clinical geneticist sense – a busy clinician couldn't care less about a genetic variant that predisposes you to wake up an hour earlier than average or have 10 fewer IQ points, or honestly even one that leads to slow-growing kidney cysts. But you might care.
Over the past few years there has been interesting activity on the 'frontiers' of molecular biology that will start to cross over to the mainstream. These movements are now starting to allow individuals to make decisions and take action based on their genetics.
1. Choosing variants to pass on to your children. Many people think about their genetics at a high level when having a baby. Because of advances in low-DNA-input genome sequencing, it's now possible to systematically test the genetic profile of embryos without destroying them. This allows couples undergoing in vitro fertilization to select which ones to implant based on the genetics of each one. So even if there's nothing you can do about your sleep patterns, IQ, or kidney cysts, it's now possible to decide which of these unique traits you'd like to pass on to the next generation.
Of course in vitro fertilization is not the norm today for most couples for a whole host of reasons, but things can change quickly. It also seems plausible that one could implement selection at the level of sperm and avoid the pain and expense of IVF for some sets of variants that are inherited from the male side.
2. Gene editing/direct interventions, or non-traditional investment into these interventions. As trust in the overall medical/research establishment has declined since 2020, there's been an interesting resurgence in self-experimentation. In a recent New Yorker article on aging, a startup CEO was memorably quoted as saying, "We don't work in mice. We work in billionaires." Outside of billionaires, communities of people with hypothyroidism started in large numbers to take the non-standard treatment of dessicated thyroid extract, and in bodybuilder forums there are threads discussing papers on new peptides to potentially impact muscle mass or fat loss.
As people with shared genetic variants find each other, two things will happen: first, the most risk-tolerant subset of people will try to experiment on themselves. If you have a loss-of-function variant in a gene, it's not too much of a stretch to imagine gene therapy or other methods to replace the missing gene. Second, the most entrepreneurial and/or wealthy subset of people will find capital to search for better interventions. For example, the 'decentralized science' community has raised millions of dollars to invest into things like longevity and long COVID research. If you know the history of how the Cystic Fibrosis Foundation partnered with Vertex to effectively cure a genetic disease, you know that pairing a motivated group of carriers of a genetic variant with capital and talent can result in enormous outcomes.
Conclusion
In sum – even if you don't know it yet, you have a set of genetic variants that make you unique, and the tools now exist such that you can choose which traits to pass to the next generation and potentially alter them in yourself.